The National Center for Microscopy and Imaging Research, a National Institutes of Health (NIH) /National Center for Research Resources (NCRR) sponsored Biomedical Technology Resource Center, is pleased to offer for download computational applications that have been developed and continue to be refined through core and collaborative research at the Center. Available tools and utilities include software for the visualization of imaging data, file format conversion tools, computational software for the reconstruction and analysis of data, software and plug-ins for the control and automation of high performance light and electron microscopes and accessories, and utilities and software to access and utilize database and data management resources. NCMIR software can be obtained without Scientific Advisory Board approval. If you have questions or need additional information call (858) 534-0276.
Visualization
Jinx
Download
- Author: Stephan Lamont
- Release Date:
- Programming Language: Java Net Beans
- Operating System:
- Other notes: Use ant to build
|
Requirements:
- Java 1.5 or more recent
- Java Web Start (installed in most modern computers)
|
Jinx is a tool for segmentation of 3D data sets. It was designed specifically for 3D electron microscopic data sets produced using electron tomography, but may be used for any aligned 3d data set. It is written in Java as a Java Webstart application. Jinx contains the facility for annotating 3D microscopic data using formal ontologies. The current version of Jinx allows the choice of ontologies, including the
Neuroscience Information Framework (NIF) ontologies, which includes an extensive ontology for subcellular anatomy of the nervous system based on our
Subcellular Anatomy Ontology (SAO)

SLASH 3D Model Viewer
Download
- Author:Jeff Bush
- Release Date:April 2, 2013
- Programming Language: WebGL
- Operating System: Windows, Linux, Mac
- Other notes: source
|
Requirements:
|
The SLASH 3D Model Viewer is a set of browser-based tools for interactive 3D displays of segmented models in a web browser. It is being designed both for in-progress models along with finalized models.

WinSage
Download
- Author: Raj Singh
- Release Date:
- Programming Language: C++
- Operating System:
- Other notes: gcc/Visual studio
|
Requirements:
|
Winsage is a Microsoft Windows compatible port (for 32-bit architectures) of the Scalable Adaptive Graphics Environment (SAGE) originally developed at the Electronic Visualization Laboratory at the University of Illinois at Chicago [http://www.evl.uic.edu/cavern/sage/].
SAGE is a graphics streaming architecture for supporting high-resolution, scalable and collaborative scientific visualization environments with potentially hundreds of megapixel of contiguous display resolution. It is primarily designed to be run as a thin middleware on cluster driven tile-displays. It allows users to treat the high-resolution distributed displays as one contiguous desktop where users can move/resize application windows. SAGE is network centric and the applications running on these displays need not run locally. The applications can be run on remote machines or clusters and can stream their pixel framebuffers to SAGE enabled tile-displays.
Winsage was developed at the National Center for Microscopy and Imaging Research under a grant by Microsoft Research. The software has been tested on Windows XP based tile-displays driven by a high-bandwidth backplane (1Gpbs) and bundles in SAGE application binaries/source for remote presentation, large 2D/3D dataset visualization and HD video playback and streaming amongst others.
Data Reconstruction & Analysis

Cytoseg
Download
- Author: Rick Giuly
- Release Date: Sept 17, 2009 (Pre-Alpha)
- Programming Language: Python
- Operating System: Windows, Linux
- Other notes: source
|
Requirements:
- Memory requirements depend on the data size
|
An open source project for automatic segmentation of neuropil. The goal of the Cytoseg project is to build tools that reduce the manual labor involved with segmentation of nueropil. Modules under development include automatic contour detectors for mitochondrion, vesicle, and cytosol regions. The project is written in Python and uses the pythonxy platform (which includes scipy and ITK image processing tools). Cytoseg is currently in a pre-alpha stage.

SLASH Online Segmentation
Download
- Author:Willy Wong
- Release Date:April 2, 2013
- Programming Language: Java
- Operating System: Windows, Linux, Mac
- Other notes: source
|
Requirements:
|
The SLASH Online Segmentation site is an open source project to enable crowdsourced and semi-automatic segmentation of massive 3D datasets, especially light and electron microscopic datasets in the field of biology.

SLASH IMOD Plugins
Download
- Author:Andrew Noske
- Release Date:April 2, 2013
- Programming Language: C
- Operating System: Windows, Linux
- Other notes: source
|
Requirements:
- Memory requirements depend on the data size
|
Bead Helper Plugin: This plugin is designed to assist in the speedy refinement of fiducial models - allowing the user to quickly identify and correct bad points.
Drawing Tools Plugin: This plugin provides a set of drawing tools which allows you to rapidly draw, modify and erase contours using the 3dmod interface in IMOD.
Interpolator Plugin: This plugin allows for rapid segmentation of surfaces by automatically generating "interpolated" contours between the "key" contours which you have manually drawn.
Livewire: Livewire, also known as Intelligent Scissors , is a segmentation technique which allows a user to select regions of interest to be extracted quickly and accurately using a relatively small number of mouse clicks.
Stereology Plugin: The IMOD Stereology plugin provides an interface for performing rapid stereology on tomographic volumes and/or stacks of images.
Image J Mosaic Plugins
Download
- Authors: Sunny Chow & Hiroyuki Hakozaki
- Release Date: 6/27/2006 (Ver. 1.1)
- Programming Language: Java
- Operating System: Support Windows platform. Tested on Windows XP and Windows Vista.
- Other notes: Used Ant to build plugin
|
Requirements:
- Mimimum Memory: 512 MByte
- Recommended Memory:1 GByte
- Storage: 200MByte
|
This package contains plugins for ImageJ for the assembly of large-scale mosaic images from a collection of overlapping image "tiles." These tiles are produced via synchronized acquisition of images as the sample is rastered over a large area utilizing motorized 3D stages. This plugin package supports manual image offset alignment, automated image offset calculation, image normalization, combining images based on alignment information, and merging three channel RGB images. Although this module is written in Java, there is a Microsoft Windows platform dependency. We currently only support this package on Windows XP Professional and Windows Vista.
Details of the image processing strategy is published in following paper: Automated microscopy system for mosaic acquisition and processing. S. K. CHOW, H. HAKOZAKI, D. L. PRICE, N. A. B. MACLEAN, T. J. DEERINCK, J. C. BOUWER, M. E. MARTONE, S. T. PELTIER & M. H. ELLISMAN, Journal of Microscopy Volume 222 Issue 2, Pages 76 – 84.
Transformation Services
Download
- Author: Asif Memon
- Version: 0.6
- Programming Language:
- Operating System: Windows/Linux/MAC
- Other notes: Use ant to build
|
Requirements:
- Java 1.4 or newer release
|
This collection of web services supports access to several online digital atlases. In the current version, the atlases include the Allen Brain Atlas, the Edinburgh Mouse Atlas Project (EMAP), brain images available registered in the UCSD spatial registry and CCDB, and the WHS (Waxholm Space) reference atlas. Each service contains a set of spatial translations that support coordinate transformations between the WHS space and the coordinate spaces used by individual atlases (including conversions between WHS and ABA voxel space, ABA reference plates, AGEA, and Paxinos/Watson reference plates). Additionally, each service can expose atlas-specific functionality. For example, the service exposing functionality of the Allen Brain Atlas would contain such requests as GetCorrelationMapByPOI, Get2dImageByPOI, GetGenesByPOI, each of them taking coordinates of a point of interest as input.
Each service supports a GetCapabilities and DescribeProcess request which can be used to describe the functionality of the service. The services are designed to be called remotely, and are currently called from a 3D atlas integration client developed at UCSD. The services are a component of the digital atlasing infrastructure (DAI) being promoted by INCF, and were initially developed to support an INCF-DAI demonstration at Neuroinformatics'2009.
TxBr
Download
- Authors: Sébastien Phan, Alex Kulongowski, Raj Singh, Masako Terada, James Obayashi and Albert Lawrence
- Version: 3.0 beta
- Programming Language: Python, C.
- Operating System: Linux, MacOsX.
- Other notes: gcc
|
Requirements:
- EPD, GiNaC, openCV, IMOD.
|
TxBR is an advanced Electron Microscopic Tomography reconstruction software that has been developed to account for the curvilinear nature of electron trajectories, making it suited for the processing of wide field Electron Microscope data. It features enhanced capabilities for performing 3D reconstruction from various schemes of data acquisition, notably multiple tilt series (any number). Versions of the code are also available to accelerate computation of massive data on clusters of CPU and GPU resources.
At this time, TxBR does not provide any tracking capabilities. However, micrograph alignment, filtering and backprojection can be done with minimal user interaction. TxBR offers additional features such as image/reconstruction dewarping and flattening that allow for high-quality montaging and Z-stacking of reconstructions. Wiki information on the scientific basis of the code may be found on the
NBCR web site.
Convertilizer
Download
- Author: Stephan Lamont
- Release Date:
- Programming Language: Java Net Beans
- Operating System:
- Other notes: Use ant to build
|
Requirements:
- Java 1.5 or more recent
- Java Web Start (installed in most modern computers)
|
Convertilizer is a tool for converting between sundry volume and image formats commonly used at NCMIR, including IMOD, Suprim, Analyze 7.5, and TIFF. It can produce image stacks, where the format permits, or take apart an image stack into separate files. It also has a facility for making Quicktime (.MOV) movies from stacks or series of images.
Instrumentation Control & Automation
Serial EM
Download
- Author: Tomas Molina and Yukiko Hakozaki
- Release Date: Summer 09
- Programming Language: C++/MFC
- Operating System: Windows only
- Other notes:
|
Requirements:
- Windows Operating System.
|
This package contains NCMIR developed plugins to the open-source SerialEM software developed and distributed by The Boulder Laboratory For 3-D Electron Microscopy of Cells, Boulder Colorado. SerialEM is a Windows based program used to acquire electron microscopic tomography (tilt-series) data on FEI and JEOL microscopes. These custom plugins enable the integration and automated control of NCMIR fielded OEM and custom digital imaging detectors (TVIPS, Gatan Inc., Spectral Instruments, Direct Electron) with SerialEM. These plugins further integrate custom control features of NCMIR’s transmission electron microscopes (e.g., stages, specimen holders, etc.) with SerialEM.
Telemicroscopy
Download
- Author: Tomas Molina
- Release Date: 2008
- Programming Language: C#
- Operating System:
- Other notes: Documentation in Fogbugs and xwiki
|
Requirements:
|
A .NET based software application was developed to remotely control NCMIR’s Jeol 3200 energy filtered electron microscope. COM packages were written to “wrap” instrument and imaging control around the microscope so as to easily expose control as simple interfaces within a C# application. Users can manipulate such parameters as magnification, brightness, focus, and condenser beam shifts. Users can also manipulate the goniometer of the electron microscope by changing such parameters as x,y,z, and tilting the specimen from \-60 to \+60 degrees. Real time video of the screen image of the microscope is provided along with a 4k x 4k high resolution image provided by our CCD cameras. Users are able to mouse click on various locations of the x,y stage and can quickly navigate large areas for surveying the specimen of study. The .NET Telemicroscopy C# application was demonstrated at SuperComputing.
Database & Data Management

CCDB Portlets
Download
- Author: Willy Wong
- Release Date:
- Programming Language: Java
- Operating System:
- Other notes: Use ant to build, GridSphere
|
Requirements:
|
The MyCCDB portal is community software for managing and sharing high resolution light and electron microscopic data. The CCDB divides the process of data entry into different stages, each with its own set of data input forms.
1) Microscopy Product Registration form assigns a unique ID to each “microscopy product”. The microscopy product is used to describe a set of related images. These images may be related in a systematic way, e.g., tilt series, optical section series, serial section, time series, mosaic, or they may be a set of survey images that were taken of the same specimen at the same time. The microscopy product ID serves as the accession number for a data set. Microscopy product ID’s can be obtained before any imaging data is acquired. Because the ID is unique, any data products labeled with this ID, e.g., negatives, can be associated with the appropriate set of details.
2) This Detailed Data input form handles the data input for more detailed data of a microscopy product, such as imaging information reconstruction and segmentation data... and it also provides users with the ability to upload data files for the datasets. Users will be able to select a pre-registered microscopy product and create a detailed dataset from the existing microscopy product.
3) The detailed Specimen Preparationform can be used to define a structured protocol recording the steps used to prepare a specimen in the order in which they were performed. It is also required for assigning a molecular localization or stain to a particular channel for light microscopy. The specimen preparation forms are still in beta testing, i.e., they are not considered production level, but should be used in order to provide feedback for the final production release.
4) The CCDB upload Tool allows users to upload multiple types of files at once into multiple forms, rather than having to upload each file individually. The tool also will recognize file types if they are named according to the CCDB naming convention and place them in the correct fields.
If your laboratory needs a data management tool for the light and electron microscopic data, the MyCCDB portal is a open source software for you to use or to extend.

CCDB Schema
Download
- Author: Willy Wong
- Release Date:
- Programming Language:
- Operating System:
- Other notes:
|
Requirements:
|
The current schema contains over 100 tables which include experimental, imaging, and reconstruction details as well as the results of any analysis of morphological and protein localization data. An overview of the schema organization is shown. All steps of 3D reconstruction are modeled in the database, from specimen preparation to the final analysis. The images are stored separately from the descriptive data, using the Storage Resource Broker for storage and retrieval of the image data sets. Oracle was chosen as a platform because its Object Relational modeling capability allows us to program any analysis or data comparison methods into the database itself. This functionality allows us to move beyond simple retrieval queries to allow us, for example, to create a “compare two protein distributions” function that runs as an integral method from within the database.
2D Image Server
Download
- Author: Asif Memon
- Release Date:
- Programming Language:Java
- Operating System:
- Other notes:Use ant to build, ArcIMS
|
Requirements:
|
This software enables client applications to communicate with remote map servers that serve 2D brain data. Currently, the servers include ESRI ArcIMS Server and the open source Minnesota MapServer. The Java wrapper supports requesting images or image fragments from these servers, taking as input the image server address, respective service name, and image fragment coordinates. The coordinates are expected to follow a coordinate system of the image as recorded in spatial registry. It is also expected that the image is warped to a canonical reference space, and registered to one of pre-defined coordinate systems (e.g. ABA reference plates, Paxinos and Watson reference plates). The software is downloadable as a single jar file. It is intended to be incorporated in a client application. In addition, both the spatial registry and servers are exposed as web services.
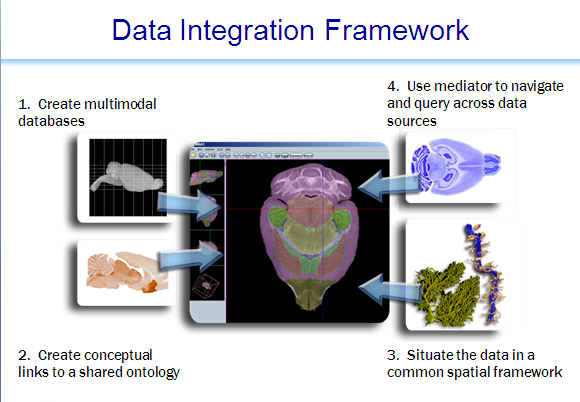
Mediator
Download
- Author: Vadim Astakhov
- Release Date:
- Programming Language: Java
- Operating System:
- Other notes:
|
Requirements:
|
System consists of a data integration server; an API for schema registration, view definition and query building;clients for registration, view definition and query composition; clients for semantic data mapping and query; a number of domain-specific clients that have incorporated access to the data integration environment (e.g. the Mouse BIRN Atlas Tool); and a set of tree\- and graph-structured ontologies and ontology-data mappings. The latter provides the necessary semantic information, tied to external ontologies, so that integrated queries can be executed across the sources.In the current global as view (GAV) setting of the system, the burden of creating proper integrated views over data sources is on the integration specialist who works with the domain scientists to capture the requirements of the application at hand. This often leads to the pragmatic problem that the relationships between attributes disclosed by different sources and between data values are, quite often, not obvious. To account for this, the system has created additional mapping relations. Currently there are three kinds of mapping relations.
1) The ontology-map relation that maps data-values from a source to a term of a recognized ontology.
2) A joinable relation links attributes from different relations if their data types and semantic types match. We have also developed a Concept Mapping client to assist in the creation of mapping entries in the term-index-source (TIS) at the time of source registration. The data integration environment has a specific source called the term-index-source that maintains a mapping between data from registered sources and ontological terms defined in various ontologies.
3) The value-maprelation maps a Mediator-supported data value or a Mediator-supported attribute-value pair to the equivalent attribute-value pair disclosed by the source.
For example, the Mediator may disclose a demographic attribute called "gender" with values (male, female), while a source may refer to "sex" with values (0, 1), while another source, with its own encoding scheme may call it "kcr_s57" with values (m, f). This is used by the system to preprocess queries.
TIS is a relational database and programmatic API which provide mechanism for mapping distributed data to standard ontological concepts.















